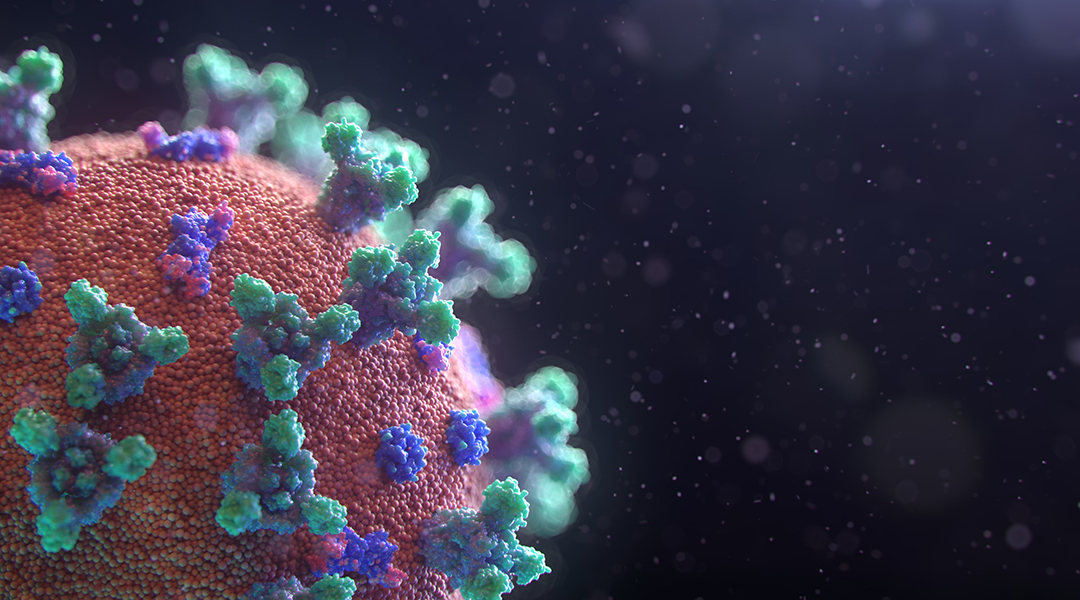
The rapidly spreading COVID-19 pandemic caused by the severe acute respiratory syndrome coronavirus (SARS-CoV-2) is on the path to become the most severe worldwide outbreak of an infectious disease in recent history, behind only the 1918 flu pandemic and HIV/AIDS.
At the time of this writing in early April 2020, over 1.5 million cases of COVID-19 have been confirmed globally, leading to a death toll exceeding 90,000, and case trajectories continuing to grow exponentially in many countries. Scientists and doctors around the world working at research institutions, hospitals and companies are pushing in a concerted effort to find drugs and develop vaccines to treat COVID-19, and to stem future outbreaks.
While a vaccine will be the preferred option to protect humanity from SARS-CoV-2, drugs to treat already infected patients or provide preventive intervention for exposed individuals are important in defending against COVID-19 mortality. Intense efforts are underway to find SARS-CoV-2 therapeutics among existing antivirals and re-purposed drugs already approved for other indications. Discovery projects have started to identify inhibitors targeting SARS-CoV-2 key proteins including the enzymes that replicate and process the viral genome.
While preventive vaccines and drugs that target viral proteins are proven routes to defend against many viral infections, RNA elements that are found in the genomes of numerous representatives within the same virus family provide new opportunities to expand the repertoire of targets for the development of antiviral therapy.
Genome expression in RNA viruses involves small regions of the genome that fold into structured RNA motifs which are often highly conserved across related viruses. These structured RNA elements play key roles in the regulation of gene expression levels and timing with regard to the viral infection cycle within host cells. Such regulatory RNA motifs function primarily as structural rather than coding components of the replication process.
Back in 2016, I discussed strategies for the discovery of drugs targeting structured RNA motifs in viruses, including an element forming an “RNA pseudoknot” in the genome of SARS-CoV. This virus, which is closely related to SARS-CoV-2, caused an outbreak of severe respiratory disease 2002-2004, albeit at a much smaller scale than the current COVID-19 pandemic.
The RNA pseudoknot motif, whose sequence is 99% conserved between the SARS-CoV and the coronavirus that causes COVID-19, folds into a convoluted knot-like structure that poses a temporary roadblock for the protein synthesis machinery of the host cell. Protein synthesis in cells is performed by ribosomes, large biomolecular complexes that translate genetic code into proteins as they slide along messenger RNA. In SARS-CoV-infected cells, ribosomes are hijacked on the viral RNA genome which directly serves as a rogue message to produce viral proteins.
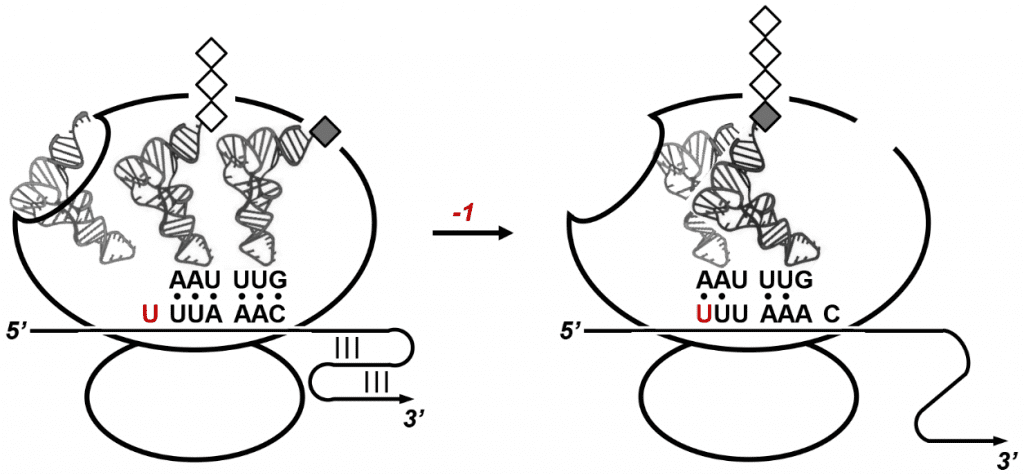
When ribosomes progressing along the SARS-CoV genome stall at the RNA pseudoknot motif, they pause over a slippery sequence immediately upstream of the pseudoknot. Ribosomes resting on the slippery sequence occasionally slide back by one nucleotide position which leads to a -1 frameshift on the viral genome. This intentional “programmed” frameshifting event enables the expression of viral enzymes required for replicating the SARS-CoV genome. Programmed ribosomal frameshifting is an evolutionary strategy to maximize the information content of compact viral genomes.
Experimental evidence from retroviruses such as HIV suggests that even small changes in frameshifting efficiency will be detrimental to virus propagation.
The frameshifting RNA pseudoknot element in the genomes of SARS coronaviruses adopts a unique structure that provides a target for the selective binding of small molecule ligands. Such ligands may affect viral protein synthesis by changing the regulatory efficiency of frameshifting events. To date, molecules that selectively bind to the SARS-CoV RNA pseudoknot and thereby inhibit viral propagation are yet elusive. Discovery efforts to identify such ligands will not deliver drugs in time to affect the current COVID-19 pandemic. However, the near-perfect conservation of the pseudoknot motif between SARS coronaviruses suggests that ligands targeting this RNA motif in the viral genome may hold a promise for the development of broad-spectrum inhibitors of SARS coronaviruses as pathogens in recurring and new future pandemics.
Written by: Thomas Hermann, Professor of Chemistry and Biochemistry and Co-Director, UCSD Center for Drug Discovery Innovation
Reference: Thomas Hermann, ‘Small molecules targeting viral RNA‘, WIREs RNA (2016). DOI: 10.1002/wrna.1373