Open-source software rapidly processes spectral data, accurately identifies and quantifies lipid species
Lipids play a key role in many metabolic diseases, including hypertension, diabetes, and stroke. So having a complete profile of the body's lipids—its "lipidome"—is important.
Lipidomics studies are often based on liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). But researchers have a hard time processing data fast enough, and they are unable to confidently identify and accurately quantify the lipid species detected.
Incorrect identifications can result in misleading biological interpretations. Yet existing tools are not designed for high-volume verification of identifications and have to be manually verified to ensure accuracy.Since scientists increasingly want larger scale lipidomics studies, analysts need improved software for identifying lipids.
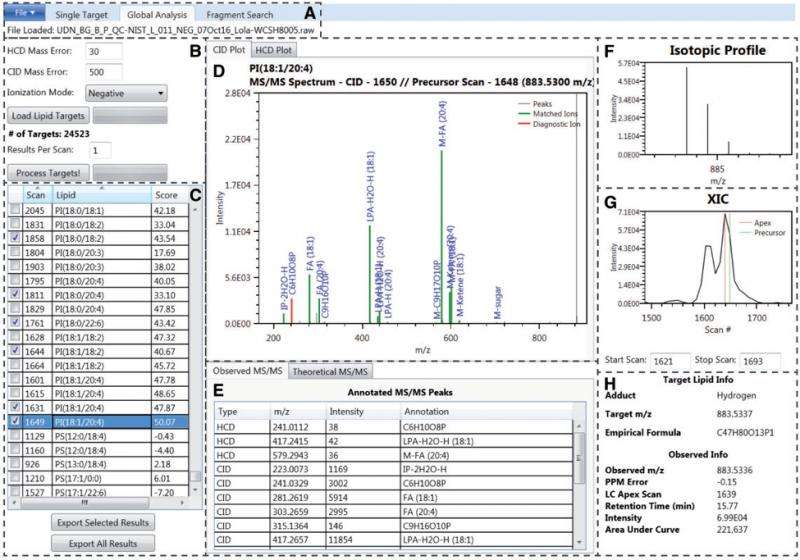
A recent paper by lead author Jennifer E. Kyle and eight co-authors at Pacific Northwest National Laboratory (PNNL) introduces an open-source lipid identification software, Lipid Quantification and Identification (LIQUID). The scoring is trainable, the search database is customizable, and multiple lines of evidence are displayed, allowing for confident identifications. LIQUID also makes single- and global-target searches available, as well as fragment-pattern searches. All this makes it possible for researchers to track similar and repeating patterns of MS/MS spectra.
Compared to other freely available software commonly used to identify lipids and other small molecules, LIQUID has a rapid processing time that can generate a higher number of validated lipid identifications faster. Its reference database includes more than 21,200 unique lipid targets across six lipid categories, 24 classes, and 63 subclasses.
LIQUID is able to confidently identify more lipid species with a faster combined processing and validation time than any other software in its field.
What's Next?
Developers of LIQUID will increase the reference library to include lipids that may be unique to particular disease states or to organisms from select environmental niches. This means researchers will be able to characterize a more diverse range of samples and therefore enhance the understanding of biological and environmental systems of interest.
More information: Jennifer E. Kyle et al. LIQUID: an-open source software for identifying lipids in LC-MS/MS-based lipidomics data, Bioinformatics (2017). DOI: 10.1093/bioinformatics/btx046
Journal information: Bioinformatics
Provided by Pacific Northwest National Laboratory